Fu, D., Calvo, J.A., Samson, L.D., 2012. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat Rev Cancer 12, 104–120. doi:10.1038/nrc3185 |
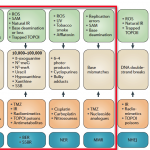
| Sources of DNA damage and their repair. Endogenous and environmental sources of DNA damage are shown in green boxes, with the lesions they cause in beige boxes (where known, the approximate number of the indicated type of lesion that occurs naturally in a cell each day is shown). Therapeutic DNA-damaging agents that cause the corresponding DNA lesion are shown in orange boxes. DNA repair pathways (blue boxes) repair DNA damage that is induced by endogenous and environmental DNA-damaging agents and thus protect the genome but they antagonize the efficacy of therapeutic DNA-damaging agents (except for mismatch repair (MMR)). BER, base excision repair; HRR, homologous recombination repair; ICL, interstrand crosslink; IR, ionizing radiation; MMC, mitomycin C; NER, nucleotide excision repair; NHEJ, non-homologous end joining; ROS, reactive oxygen species; SAM, S‑adenosyl methionine; SSB, single-strand break; SSBR, SSB repair; TMZ, temozolomide; TOPO, topoisomerase; UV, ultraviolet |
| A chart of common DNA damaging agents, examples of lesions they cause in DNA, and pathways used to repair these lesions. Also shown are many of the genes in these pathways, an indication of which genes are epigenetically regulated to have reduced (or increased) expression in various cancers. It also shows genes in the error prone microhomology-mediated end joining pathway with increased expression in various cancers. |
Single-strand damage (PARP depending)
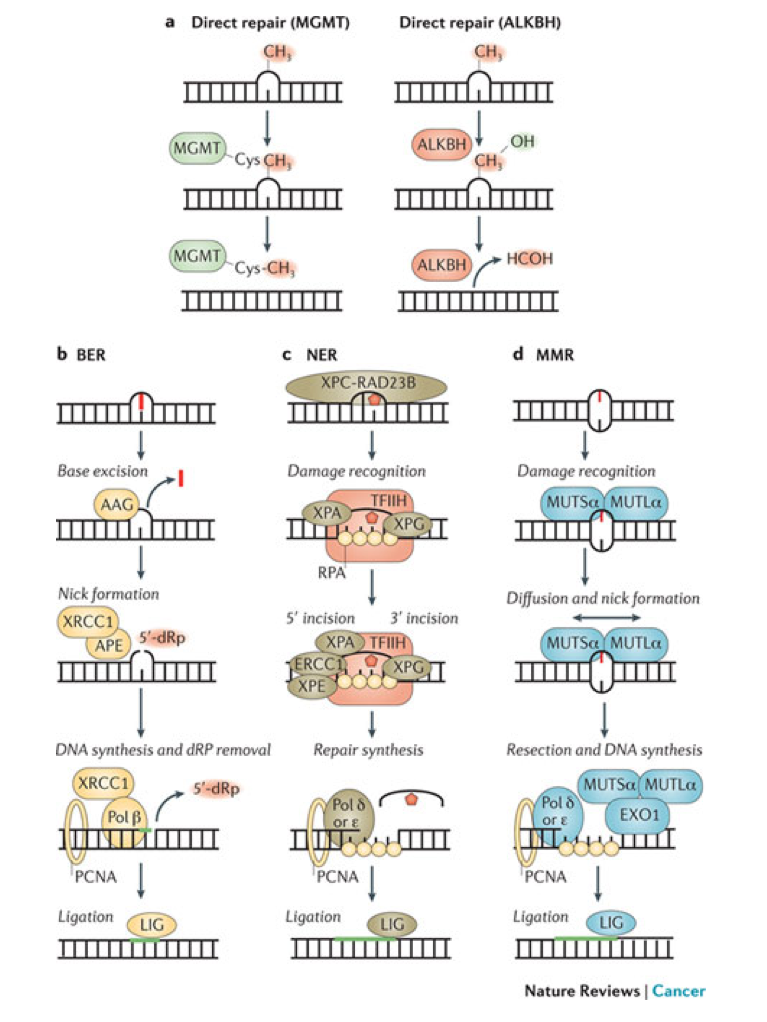 |
a
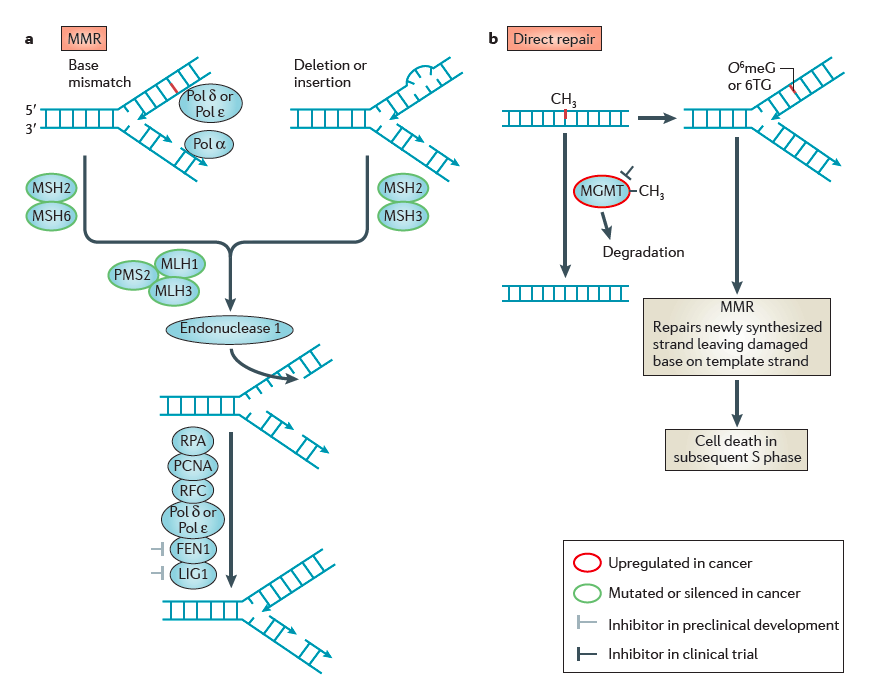
- | Direct repair results in the reversal of an alkylated base to a normal base without any intervening steps or the generation of DNA strand breaks. The O 6–MethylGuanine DNA MethylTransferase (MGMT) catalyses the transfer of the aberrant methyl group onto a cysteine in the active site of MGMT, thereby removing the lesion from DNA. The AlkB homologue (ALKBH) family of α-ketoglutarate-dependent dioxygenases catalyse the hydroxylation of the aberrant methyl group, which then spontaneously leaves as formaldehyde (HCOH) to regenerate the normal base.
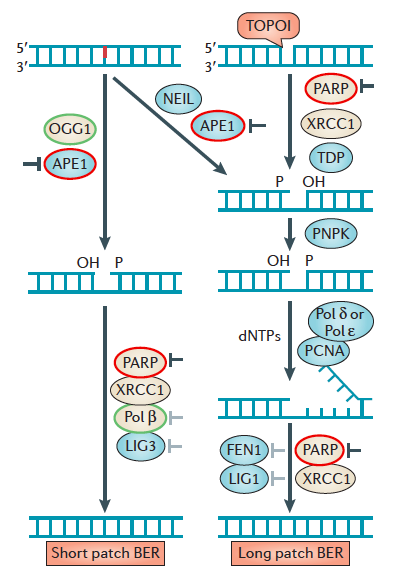
- The base excision repair (BER) pathway removes simple alkyl and oxidative base lesions. BER is initiated by a DNA glycosylase, such as alkyladenine-DNA glycosylase (AAG), which excises the damaged base to generate an abasic site for subsequent processing by either short-patch BER or long-patch BER (not shown). In short-patch BER, the apurinic-apyrimidinic endonuclease (APE) incises the abasic site to yield a 3′ OH adjacent to a 5′-deoxyribose phosphate (5′-dRP) moiety. DNA polymerase β (Pol β) removes the 5′-dRP and replicates DNA from the 3′ OH, generating a nick ready for subsequent ligation by DNA ligase (LIG). Long-patch repair occurs in a similar manner except that a longer piece of DNA is removed.
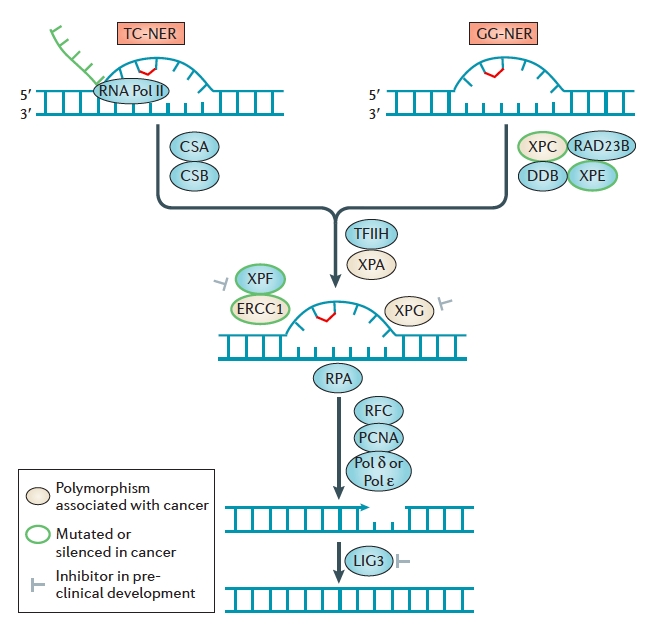
- Nucleotide excision repair (NER) removes bulky, helix-distorting lesions but it can also serve as a backup repair mechanism for the excision of certain alkylated bases16. NER is initiated by the recognition of a distorted region containing the DNA adducts followed by a 5′ and 3′ incision on either side of the lesion to create a 27–29 nucleotide gap. The excised region is then filled in by Pol δ or Pol ɛ for subsequent ligation.
- The mismatch repair (MMR) pathway mediates the removal of mismatched bases, as well as mispairs that are caused by base alkylation lesions such as O 6meG:T. In mammals, MMR is initiated by the recognition of mismatches by the MUTSα heterodimer (MSH2–MSH6) followed by recruitment of the MUTLα heterodimer (PMS2–MLH1). Diffusion of MUTSα–MUTLα leads to nicking of the DNA either upstream or downstream of the mismatch; this serves as an entry point for exonuclease 1 (EXO1) that removes a segment of DNA. This is subsequently filled in and repaired by a combination of Pol δ, Pol ɛ and LIG to complete repair. TFIIH, transcription factor IIH; XP, xeroderma pigmentosum protein.
|